
- ENGAUGE DIGITIZER PUBLICATION PDF
- ENGAUGE DIGITIZER PUBLICATION SOFTWARE
- ENGAUGE DIGITIZER PUBLICATION SIMULATOR
Is there something wrong with this publication? Contact our curators. Reproducible computational biology experiments with. A quantitative description of membrane current and its application to conduction and excitation in nerve. Plot Digitizer (PD) and WebPlotDigitizer (WD) using published KM curves.
ENGAUGE DIGITIZER PUBLICATION SOFTWARE
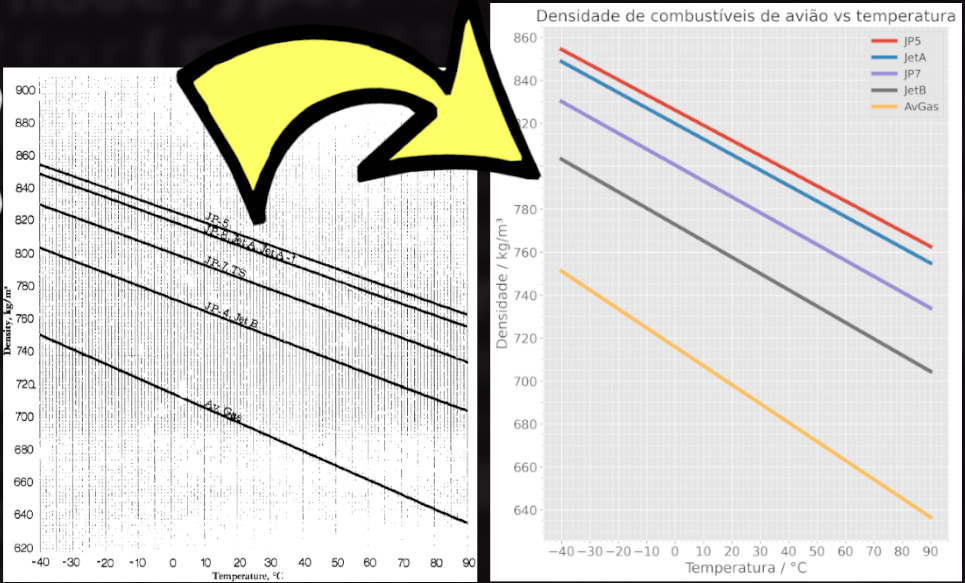
Journal: General Pharmacology: The Vascular System. A Google search identified 13 different free graph digitization software tools. Includes an automatic digitization feature that can automatically digitize many types of functional data. Using axis point and curve point tools, one can define the graph axis and data set points of an imported image, respectively. An easy to use Java program that allows you to digitize data points off of scanned plots, scaled drawings, or orthographic photographs. ten Tusscher, Kirsten HWJ and Noble, Denis and Noble, Peter-John and Panfilov, Alexander V. For data extraction from images, authors can use Engauge digitizer ( effectively and reliably to import images from an article directly or via Microsoft Paint. Journal: American Journal of Physiology-Heart and Circulatory Physiology. Poh, Yong Cheng and Corrias, Alberto and Cheng, Nicholas and Buist, Martin Lindsay. The resulting data points are usually used as input to other software applications ( ). The software accepts image files (such as PNG, JPEG, and TIFF) containing graphs and recovers the data points from those graphs. A quantitative model of human jejunal smooth muscle cell electrophysiology. The Engauge Digitizer software was employed to extract the data from figures. (v3): This Abstract was appended with a note on changes made to versions.
EDITOR'S NOTE (v2): A typographical correction in the reproducibility report was made. In addition, some inconsistencies have been uncovered and discussed in this paper. We create a modularized CellML implementation of the model, which is able to reproduce clamping behaviours of individual currents and whole cell action potential traces. The ionic currents are described by either a traditional Hodgkin-Huxley (HH) formalism or a deterministic multi-state Markov (MM) formalism.
(2012) paper describes the first biophysically based computational model of human jejunal smooth muscle cell (hJSMC) electrophysiology. We thus carried out a comprehensive system review and meta-analysis investigating the prognostic significance and clinicopathological features of SMAD4 gene mutation in CRC patients. Primary Publication: A Quantitative Model of Human Jejunal Smooth Muscle Cell Electrophysiology. Background Approximately 5.024.2 of colorectal cancers (CRCs) have inactivating mutations in SMAD4, making it one of the frequently mutated genes in CRC.
ENGAUGE DIGITIZER PUBLICATION SIMULATOR
from publication: Design of a PC-Based Patient Simulator for.
ENGAUGE DIGITIZER PUBLICATION PDF
Īll Physiome articles are published as a collection containing the manuscript as a PDF file and the model implementation as an OMEX file. Download scientific diagram ECG signal digitalisation process using Engauge Digitizer v4.12. To cite this Physiome article, cite the whole collection at the DOI: 10.36903/physiome.16590317, and the Primary Publication at the DOI. A Quantitative Model of Human Jejunal Smooth Muscle Cell Electrophysiology


 0 kommentar(er)
0 kommentar(er)
